This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Lipid nanoparticle-based ribonucleoprotein delivery for in vivo genome editing

Gene therapy is a potential mode of treatment for a wide variety of diseases caused by genetic mutations. While it has been an area of diverse and intense research, historically, only a very few patients have been treated using gene therapy—and fewer still cured. The advent of the genetic modification technique called CRISPR-Cas9 in 2012 has revolutionized gene therapy—as well as biology as a whole—and it has recently entered clinical trials for the treatment of some diseases in humans.
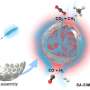
Haruno Onuma, Yusuke Sato and Hideyoshi Harashima at Hokkaido University have developed a new delivery system for CRISPR-Cas9, based on lipid nanoparticles (LNPs), that could greatly increases the efficiency of in vivo gene therapy. Their findings were published in the Journal of Controlled Release.
"There are broadly two ways of treating diseases with gene therapy," Sato explained, "ex vivo, where cells are subjected to the desired modifications in the laboratory and then introduced into the patient, and in vivo, where the treatment is administered to the patient to change the cells in their body. Safe and effective in vivo treatment is the ultimate aspiration of gene therapy, as it would be a straightforward process for patients and healthcare providers. LNPs can function as a vehicle for the safe and effective delivery of such therapies."
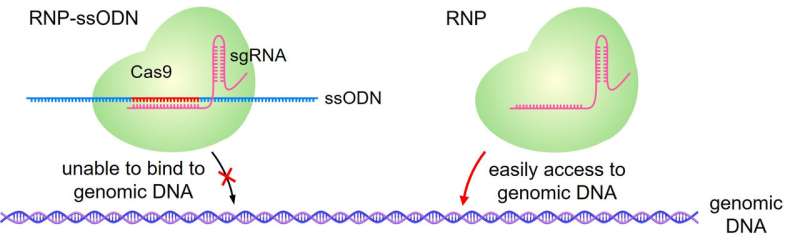
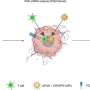
CRISPR-Cas9 consists of a large molecule composed of the Cas9 protein and guide RNA. The guide RNA binds to a specific, complementary DNA sequence, and the Cas9 protein cuts that sequence, allowing it to be modified. The guide RNA can be altered to target specific DNA sequences to be modified.
"In a previous study, we discovered that additional DNA molecules, called ssODNs, ensure that the CRISPR-Cas9 molecule is loaded into the LNPs (CRISPR-LNPs)," Harashima elucidated. "In this study, we again used ssODNs, but they were carefully designed so that they would not inhibit the function of the guide RNA."
Using a guide RNA targeting the expression of a protein called transthyretin, they evaluated the effectiveness of the CRISPR-LNPs in mice models. CRISPR-LNPs with ssODNs that dissociated from the guide RNA at room temperature were most effective at reducing serum transthyretin: two consecutive doses, one day apart, reduced it by 80%.
"We have demonstrated the optimal ssODN sequence affinity that ensures the loading and the release of CRISPR-Cas9 at the target location; and that this system can be used to edit cells in vivo," concluded Onuma. "We will continue to improve the design of ssODNs, as well as to develop optimal lipid formulations to increase the effectiveness of delivery."
More information: Haruno Onuma et al, Lipid nanoparticle-based ribonucleoprotein delivery for in vivo genome editing, Journal of Controlled Release (2023). DOI: 10.1016/j.jconrel.2023.02.008
Journal information: Journal of Controlled Release
Provided by Hokkaido University