Fast, uncomplicated, and specific: Diagnosis of prostate cancer from blood samples
Early detection of prostate cancer, one of the most common types of cancer in men, is often achieved with PSA tests. However, this blood test for prostate-specific antigens gives many false positive results, causing unnecessary biopsies and overtreatment. In the journal Angewandte Chemie International Edition, a Chinese research team now introduces a highly specific, non-invasive alternative to biopsy: the "thermophoretic AND gate operation" assay, abbreviated as Tango, quickly and reliably detects prostate cancer directly in blood samples.
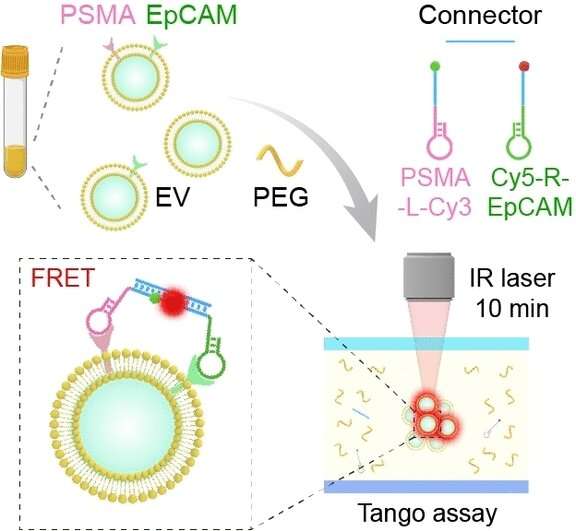
The Tango assay is based on the analysis of circulating extracellular vesicles, which are membrane-bound "nanobubbles." These come from all cells of the body, circulate in the bloodstream, and contain numerous biomarkers typical of the cells in which they originated. Isolation and accumulation of the heterogeneous vesicles in complex samples requires complex and expensive pre-treatments. The new method developed by a team headed by Fei Tian, Bo Dai, and Jiashu Sun combines accumulation with a logical AND gate operation in a single step for the identification of the desired tumor vesicles.
The concentration process is based on thermophoresis, the movement of particles based on a temperature gradient. The sample is placed into a specially designed microchamber that is locally heated with an IR laser. The vesicles preferentially move toward the heated spot. Polyethylene glycol is also added to form a concentration gradient, which amplifies the effect. This results in a 2800-fold accumulation around the laser spot.
To identify the desired vesicles unequivocally and specifically, they must contain two proteins that occur in high concentrations in prostate tumors: prostate-specific antigen (PSMA) and epithelial cell-adhesion molecule (EpCAM). The team introduced two probes based on aptamers, which are short, single strands of DNA with a "programmed" 3D structure that specifically binds to a target molecule. In this case the two targets are PSMA and EpCAM. Each of the probes has fluorescence dye.
In order to only detect vesicles that contain both tumor markers, the team developed a logical AND operation. Both of the probes have a little molecular "anchor" that specifically binds to the end of a DNA connector. If both of the target proteins are found on a vesicle membrane, both types of probe are linked by the DNA connector and the two fluorescence dyes come close enough to each other for an energy transfer. The one dye absorbs light and transfers part of the energy to the other without radiation (Förster resonance energy transfer, FRET), the second dye then emits light. The intensity of this FRET fluorescence is a measure of the number of vesicles containing both tumor markers.
The Tango assay was able to identify patients with prostate cancer out of a group with inconclusive PSA results with 91 % accuracy in 15 minutes. It should also be possible to develop tango tests for other types of cancer, according to the team from the National Center for Nanoscience and Technology (Beijing) and Fudan University (Shanghai).
More information: Jinqi Deng et al, One‐Step Thermophoretic AND Gate Operation on Extracellular Vesicles Improves Diagnosis of Prostate Cancer, Angewandte Chemie International Edition (2022). DOI: 10.1002/anie.202207037
Journal information: Angewandte Chemie International Edition , Angewandte Chemie
Provided by Angewandte Chemie International Edition