Scientists at Stockholm's KTH Royal Institute of Technology have mapped the genomes of bacterioplankton in the Baltic Sea, finding that their closest relatives are in brackish North American estuaries such as Chesapeake and Delaware bays. Credit: KTH Royal Institute of Technology
The first large-scale mapping of genomes of bacterial plankton in the Baltic Sea shows that the bacterias' closest relatives are not in oceans or freshwater lakes, but in other brackish environments. The genomes may not yet answer where these plankton came from, but they will help scientists to better understand brackish, or briny, ecosystems.
In a study published in Genome Biology, researchers from Sweden's KTH Royal Institute of Technology's SciLifeLab research center and Linnaeus University used DNA sequencing and advanced algorithms to reconstruct the genomes of a number of bacterial plankton, or bacterioplankton, in the Baltic Sea.
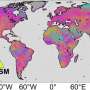
A comparison with genetic data from bodies of water around the world shows that Baltic bacterioplankton are genetically distinct from their relatives in both oceans and freshwater lakes. Several of the genomes are similar to those of another large brackish water system, Chesapeake Bay on the East Coast of North America. Andersson says this suggests that there is a global microbiome specializing in brackish water, which split off from ocean and freshwater plankton at least 100,000 years ago—long before the Baltic became a brackish sea, 8000 years ago.
But from where these bacterial plankton originate—and how they ended up in the Baltic—has yet to be determined.
Andersson says the bacterioplankton have a different history than multicellular organisms in the Baltic, which are by contrast locally adapted populations of freshwater or marine counterparts.
The findings aren't only about the relationships between plankton families—there is an environmental significance to the study, too. Since bacteria are important for the cycles of carbon and nutrients in the sea, this is an important piece of the puzzle in understanding how the Baltic Sea ecosystem works, Andersson says.
"This is a big step forward in the effort to increase understanding of how nutrients are converted in the Baltic Sea, and how these processes will be affected by global warming," Andersson says.
A litre of seawater contains about 1 billion microscopic plankton, much of which is bacteria and its cousins, archaea. These unicellular organisms belong to a bunch of different species all specialized in different tasks for their survival. Some perform, for example, photosynthesis (cyanobacteria); others feed on carbon compounds from other plankton or runoff from the land; and a few get their energy from catalyzing the oxidation of various non-organic molecules, such as ammonia.
Most marine bacterial species are difficult to cultivate. Yet with a method called metagenomics, the DNA can be sequenced directly from a water sample, and by piecing together the short DNA strands, a more or less complete genome can be obtained. The scientists took water samples weekly for one year at a station 10 km east of the island of Öland. The DNA was extracted and sequenced at SciLifeLab in Stockholm. The team used a supercomputer to piece together the short DNA strands into longer genetic fragments and eventually the researchers used an algorithm they developed for sorting the fragments into genomes of different species.
"Metagenomics gives us a window to the world of microbes. By identifying enzymes and uptake mechanisms encoded in the genomes of different plankton species, we can understand what nutrients they live on. In the long term this will lead to better models of nutrient cycling in the oceans," says lead author Luisa Hugerth, a doctoral student at KTH.
Andersson says that for environmental research in the Baltic, mapping the bacterioplankton genomes serves a similar purpose as the mapping of the human genome does for medical research, where the information about the human genome is a pillar and an absolute necessity for much of the research being conducted today. "Now that this part is finished, work is fully underway to map the genomes of the most common microscopic plankton species in the Baltic," he says.
More information: Metagenome-assembled genomes uncover a global brackish microbiome, Luisa W. Hugerth, John Larsson, Johannes Alneberg, Markus V. Lindh, Catherine Legrand, Jarone Pinhassi and Anders F. Andersson, Genome Biology 2015, 16:279 DOI: 10.1186/s13059-015-0834-7
Journal information: Genome Biology
Provided by KTH Royal Institute of Technology