Genes that express in precisely timed patterns, known as oscillatory genes, play an essential role in development functions like cell division, circadian rhythms and limb formation. But without a time-lapse view of genetic expression, these genes have gone largely undiscovered.
An algorithm developed by scientists at the Morgridge Institute for Research and the University of Wisconsin-Madison is giving scientists a new way to identify the dynamics of oscillatory genes, and perhaps defining the roles of these early-development forces for the first time.
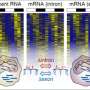
A paper published in this week's online edition of Nature Methods describes this new statistical approach, called "Oscope," which helps identify oscillating genes in single-cell RNA-sequencing experiments. The key to Oscope is examining cells from an unsynchronized population, where the cells are in different developmental states.
Studying oscillatory genes using traditional RNA-seq technology requires investigators to "pick" only one known oscillatory system and perform synchronization toward this system. This step masks the oscillatory signals of any other systems.
"We study unsynchronized single cell data here so none of the oscillatory systems are disturbed," says Ning Leng, a computational biologist with the Morgridge Institute and co-author of the paper. "This enables us to discover the unknown oscillatory signals."
Oscope identifies independent groups of cyclic genes and captures one "base cycle" of each group, offering a practical way to profile distinct groups of genes that play a cyclical role. This technology could open new research roads into genes that are at the heart of basic development.
Oscope was born out of an interdisciplinary collaboration. To solve the problem required combining the statistical expertise of Professor Christina Kendziorski's group from the Department of Biostatistics and Medical Informatics at UW-Madison with the cell biology strengths of stem cell research pioneer James Thomson's group at the Morgridge Institute for Research. Leng worked on the project as a graduate student in Kendziorski's group, and joined the Thomson lab upon graduation this fall.
Leng co-authored the paper with Li-Fang Chu, a Morgridge postdoctoral research associate. Both scientists are part of Thomson's Morgridge Regenerative Biology team. Leng began the project in 2013 as a graduate student in Kendziorski's group.
The Oscope software package is offered free to scientists at this website..
"We view this as the first step to understanding oscillatory genes and their relationship to developmental processes," says Chu. "What's most interesting is, how are the genetic regulatory networks governing oscillatory gene expression? How are these signals coordinated in a timely fashion to execute a developmental task?"
The goal of the Thomson lab will be to establish a robust system for the detection of timing-related genes that are important to developmental process, Chu says.
The most well understood oscillatory genes are those related to the circadian clock, which is based on sunlight and tightly regulates metabolism and cell proliferation over a 24-hour period.
There are disease implications as well, says Chu, the most notable being cancer, where the normal timing of cell division is hijacked as cells proliferate out of control. The Oscope technique may identify other connections between genetic cycles and disease.
Single-cell RNA sequencing is an extremely powerful tool for getting snapshots of gene expression, but is less able to trace a cell over time. Oscope uses the high-resolution snapshot data to identify different gene groups that are potentially changing over time.
"We need a combination of an average signal across many cells for the big picture, and we also need single-cell resolution techniques to compare the two and figure out which signal may represent true biological variables," Chu says. "The data analysis is very challenging and requires novel algorithms, such as Oscope, to perform these complex tasks."
More information: Oscope identifies oscillatory genes in unsynchronized single-cell RNA-seq experiments, Nature Methods (2015) DOI: 10.1038/nmeth.3549
Journal information: Nature Methods
Provided by Morgridge Institute for Research